Group highlights
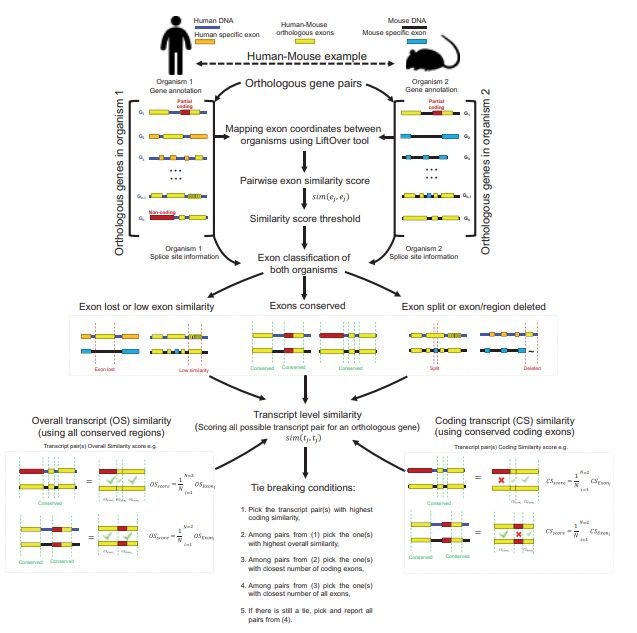
We describe a new method, ExTraMapper, which extracts fine-scale mappings between the exons and transcripts of a given pair of ortholog genes between two organisms using sequence conservation between the two genomes and their gene annotations.
Chakraborty A, Ay F, Davuluri RV
Bioinformatics 2021 May 20;37(20):3412–20
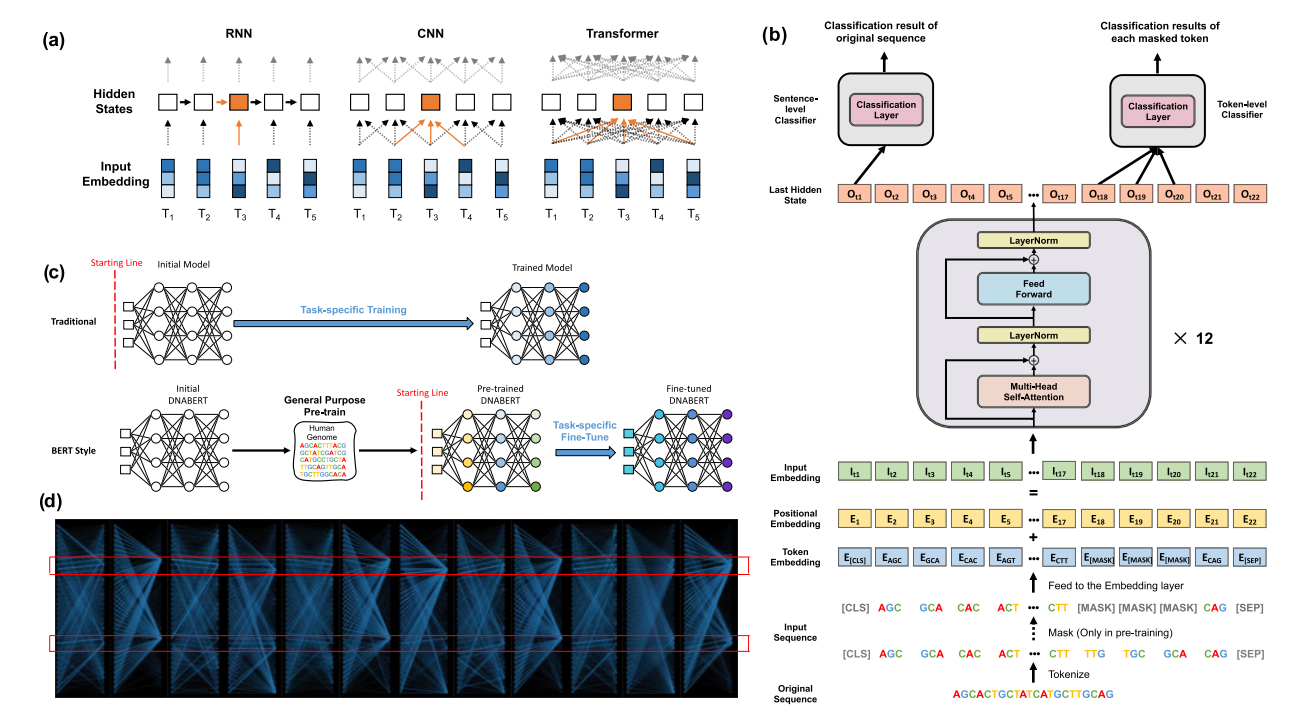
We adapted the idea of Bidirectional Encoder Representations from Transformers (BERT) model to genomic DNA setting and developed a deep learning method called DNABERT.
Ji Y, Zhou Z, Liu H, Davuluri RV
Bioinformatics 2021 Feb 4:btab083
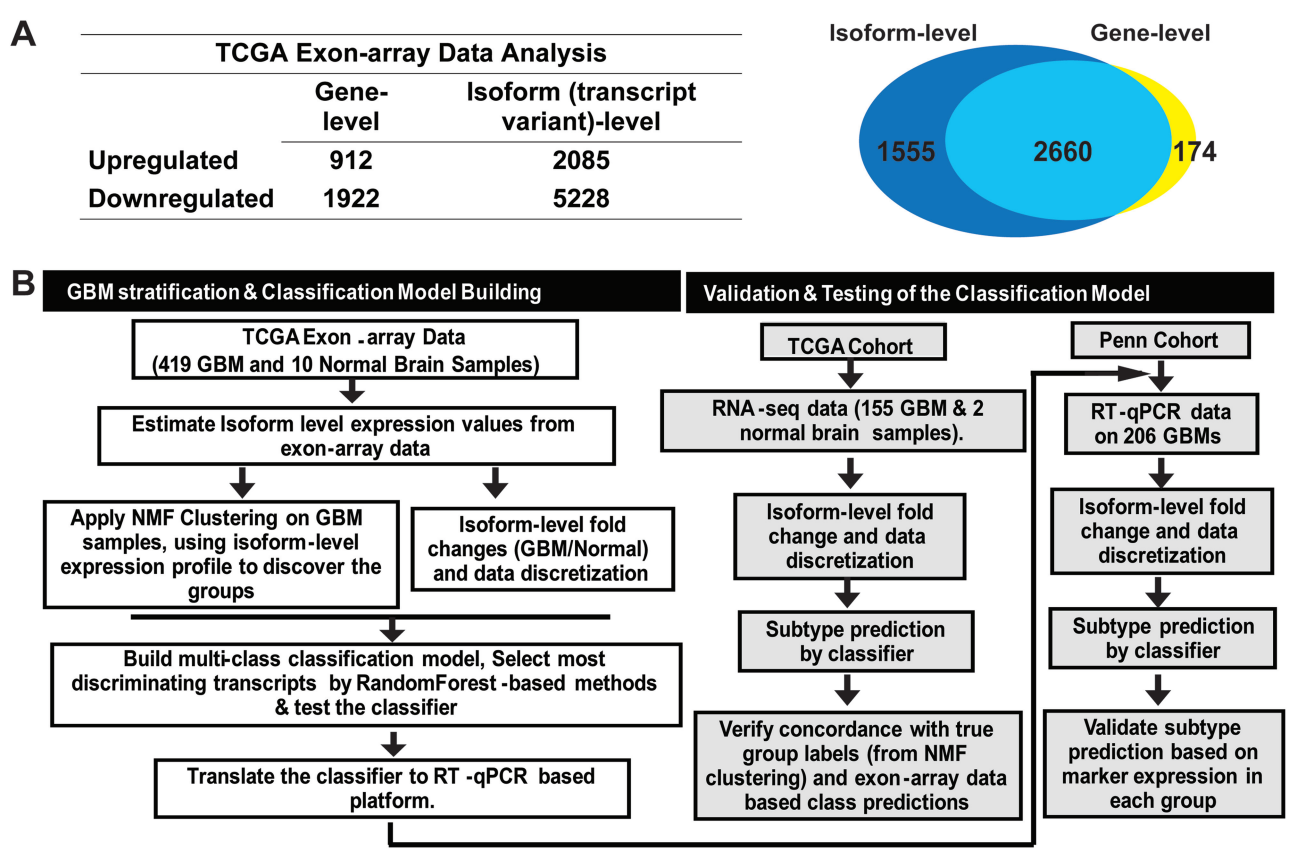
We designed a platform-independent isoform-level gene-expression based classification system (PIGExClass) that would allow us transfer the gene-signature developed on a high-dimensional platform to a clinically adaptable low-dimensional platform.
Pal S, Bi Y, Macyszyn L, Showe LC, O’Rourke DM, Davuluri RV
Nucleic Acids Res 2014 Apr;42(8):e64
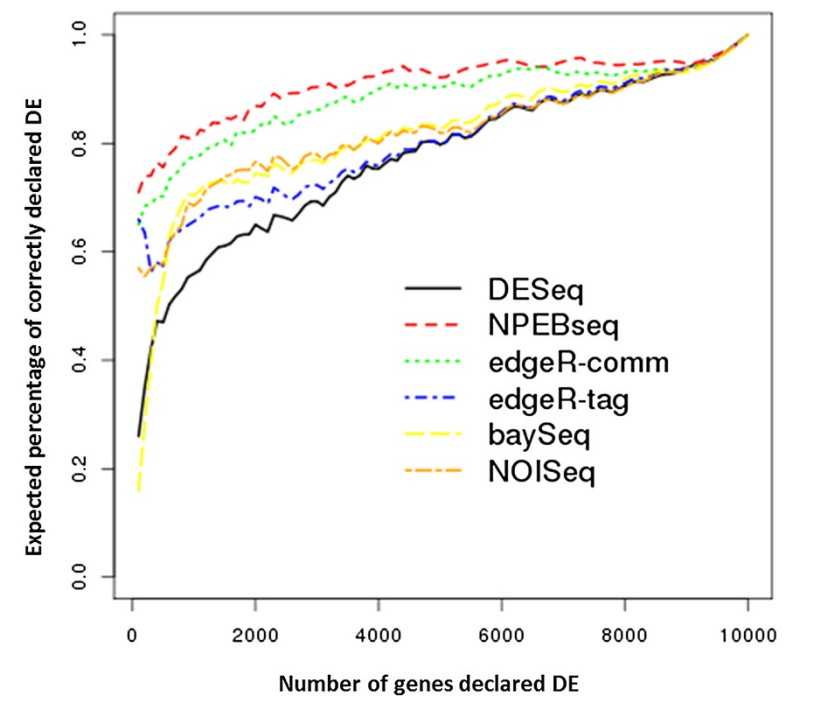
We developed a novel method to model the RNA-seq data and detect differentially expressed genes and exons across different conditions. The method is compared with the other popular methods for RNA-seq DE analysis, both using simulated and real RNA-seq datasets.
Bi Y, Davuluri RV
BMC Bioinformatics 2013 Aug 27;14:262
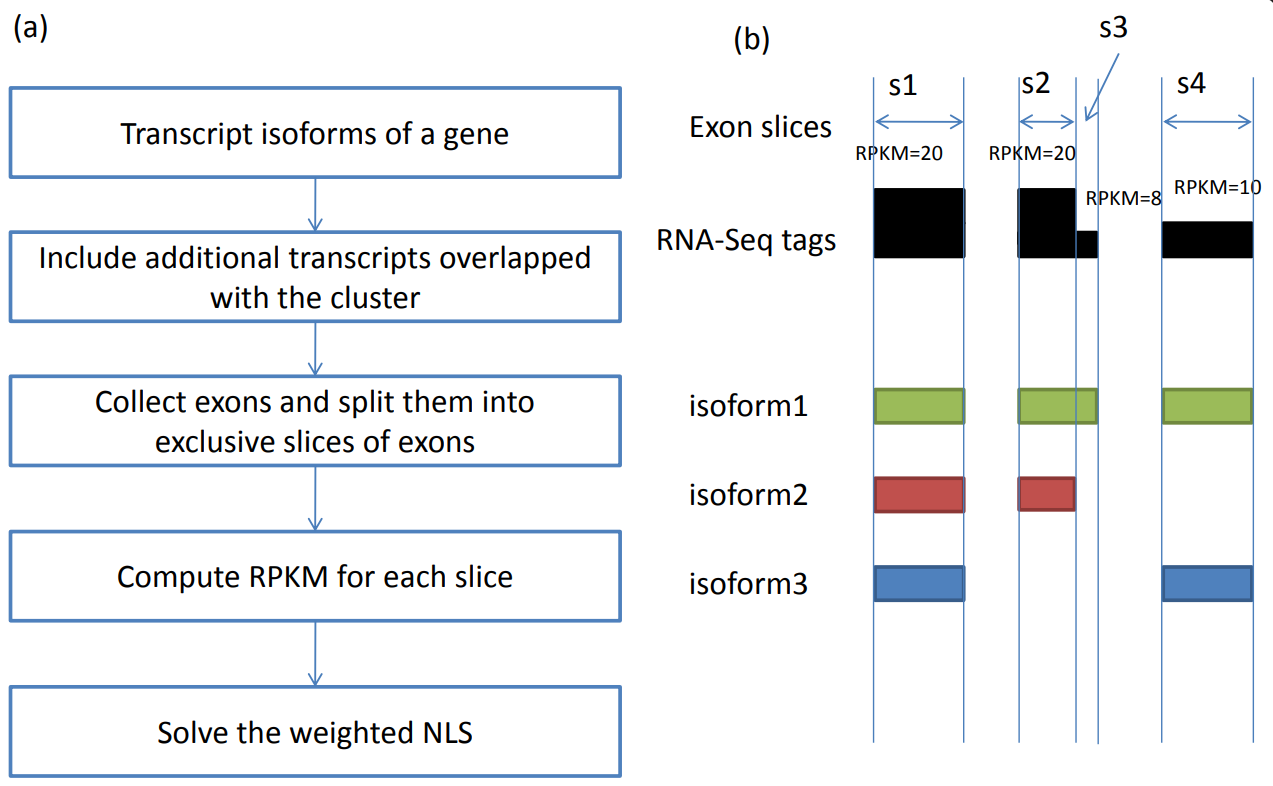
We present a novel method, called IsoformEx, which is based on weighted non-negative least squares and compared IsoformEx with some of the published methods in accurately estimating the expression levels of transcript isoforms.
Kim H, Bi Y, Pal S, Gupta R, Davuluri RV
BMC Bioinformatics 2011 Jul 27;12:305
Publications
Computational identification of promoters and first exons in the human genome
Davuluri RV, Grosse I, Zhang MQ
Nat Genet 2001 Dec;29(4):412-7
AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors
Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E
BMC Bioinformatics 2003 Jun 23;4:25
Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility
Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M, Wojcik SE, Ferdin J, Kunej T, Xiao L, Manoukian S, Secreto G, Ravagnani F, Wang X, Radice P, Croce CM, Davuluri RV, Calin GA
Cancer Res 2010 Apr 1;70(7):2789-98
The functional consequences of alternative promoter use in mammalian genomes
Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH
Trends Genet 2008 Apr;24(4):167-77
AGRIS and AtRegNet
Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E
a platform to link cis-regulatory elements and transcription factors into regulatory networks Plant Physiol
ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing
Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, Iizasa H, Davuluri RV, Nishikura K
Cell 2013 Apr 25;153(3):575-89
CART classification of human 5’ UTR sequences
Davuluri RV, Suzuki Y, Sugano S, Zhang MQ
Genome Res 2000 Nov;10(11):1807-16
Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development
Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, Dahmane N, Davuluri RV
Genome Res 2011 Aug;21(8):1260-72
Epigenetic profiling in chronic lymphocytic leukemia reveals novel methylation targets
Rush LJ, Raval A, Funchain P, Johnson AJ, Smith L, Lucas DM, Bembea M, Liu TH, Heerema NA, Rassenti L, Liyanarachchi S, Davuluri R, Byrd JC, Plass C
Cancer Res 2004 Apr 1;64(7):2424-33
Alternative transcription and alternative splicing in cancer
Pal S, Gupta R, Davuluri RV
Pharmacol Ther 2012 Dec;136(3):283-94
Identifying estrogen receptor alpha target genes using integrated computational genomics and chromatin immunoprecipitation microarray
Jin VX, Leu YW, Liyanarachchi S, Sun H, Fan M, Nephew KP, Huang TH, Davuluri RV
Nucleic Acids Res 2004 Dec 17;32(22):6627-35
Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs
Jin VX, Singer GA, Agosto-Pérez FJ, Liyanarachchi S, Davuluri RV
BMC Bioinformatics 2006 Mar 7;7:114
An integrative ChIP-chip and gene expression profiling to model SMAD regulatory modules
Qin H, Chan MW, Liyanarachchi S, Balch C, Potter D, Souriraj IJ, Cheng AS, Agosto-Perez FJ, Nikonova EV, Yan PS, Lin HJ, Nephew KP, Saltz JH, Showe LC, Huang TH, Davuluri RV
BMC Syst Biol 2009 Jul 17;3:73
FTO-Dependent N6-Methyladenosine Modifications Inhibit Ovarian Cancer Stem Cell Self-Renewal by Blocking cAMP Signaling
Huang H, Wang Y, Kandpal M, Zhao G, Cardenas H, Ji Y, Chaparala A, Tanner EJ, Chen J, Davuluri RV, Matei D
Cancer Res 2020 Aug 15;80(16):3200-3214
Immune landscapes associated with different glioblastoma molecular subtypes
Martinez-Lage M, Lynch TM, Bi Y, Cocito C, Way GP, Pal S, Haller J, Yan RE, Ziober A, Nguyen A, Kandpal M, O’Rourke DM, Greenfield JP, Greene CS, Davuluri RV, Dahmane N
Acta Neuropathol Commun 2019 Nov 29;7(1):203
Isoform-level gene signature improves prognostic stratification and accurately classifies glioblastoma subtypes
Pal S, Bi Y, Macyszyn L, Showe LC, O’Rourke DM, Davuluri RV
Nucleic Acids Res 2014 Apr;42(8):e64
Isoform level expression profiles provide better cancer signatures than gene level expression profiles
Zhang Z, Pal S, Bi Y, Tchou J, Davuluri RV
Genome Med 2013 Apr 17;5(4):33
Genome-wide analysis of alternative promoters of human genes using a custom promoter tiling array
Singer GA, Wu J, Yan P, Plass C, Huang TH, Davuluri RV
BMC Genomics 2008 Jul 25;9:349
Genome-wide mapping of RNA Pol-II promoter usage in mouse tissues by ChIP-seq
Sun H, Wu J, Wickramasinghe P, Pal S, Gupta R, Bhattacharyya A, Agosto-Perez FJ, Showe LC, Huang TH, Davuluri RV
Nucleic Acids Res 2011 Jan;39(1):190-201
ExTraMapper: Exon- and Transcript-level mappings for orthologous gene pairs
Chakraborty A, Ay F, Davuluri RV
Bioinformatics 2021 May 20;37(20):3412–20
DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome
Ji Y, Zhou Z, Liu H, Davuluri RV
Bioinformatics 2021 Feb 4:btab083
Isoform-level gene signature improves prognostic stratification and accurately classifies glioblastoma subtypes
Pal S, Bi Y, Macyszyn L, Showe LC, O’Rourke DM, Davuluri RV
Nucleic Acids Res 2014 Apr;42(8):e64
NPEBseq: nonparametric empirical bayesian-based procedure for differential expression analysis of RNA-seq data
Bi Y, Davuluri RV
BMC Bioinformatics 2013 Aug 27;14:262
IsoformEx: isoform level gene expression estimation using weighted non-negative least squares from mRNA-Seq data
Kim H, Bi Y, Pal S, Gupta R, Davuluri RV
BMC Bioinformatics 2011 Jul 27;12:305
For a full list of publications go to Google Scholar, DBLP
Patents
Methods and compositions for diagnosis of glioblastoma or a subtype thereof
Ramana V. Davuluri, Sharmistha Pal, Yingtao Bi, Louise C. Showe, Donald M. O’rourke, Luke Macyszyn
US10113201B2 (2018)
Diagnositic Methods of Tumor Susceptibility With Nucleotide Polymorphisms Inside MicroRNA Target Sites
Milena Nicoloso, George Calin, Hao Sun, Ramana Davuluri
US20120322069A1 (2012)